
Recently, the Microbial and Enzyme Engineering Science and Technology Innovation Team at the Institute of Animal Science, Chinese Academy of Agricultural Sciences (CAAS), has developed an innovative deep learning-based framework, DLFea4AMPGen, for the de novo design of multifunctional antimicrobial peptides (AMPs). This new method allows for efficient screening and generation of peptides with diverse biological activities, offering great promise in the intelligent design and development of AMPs. The results of this research were published in Nature Communications.
AMPs, which are natural defense molecules found in plants, animals, and microorganisms, have gained significant attention as potential alternatives to antibiotics. They can physically disrupt bacterial membranes, bypassing existing resistance mechanisms. However, current de novo design strategies for AMPs still face challenges, such as low success rates and the need for large virtual peptide libraries, which hinder the speed and feasibility of their development.
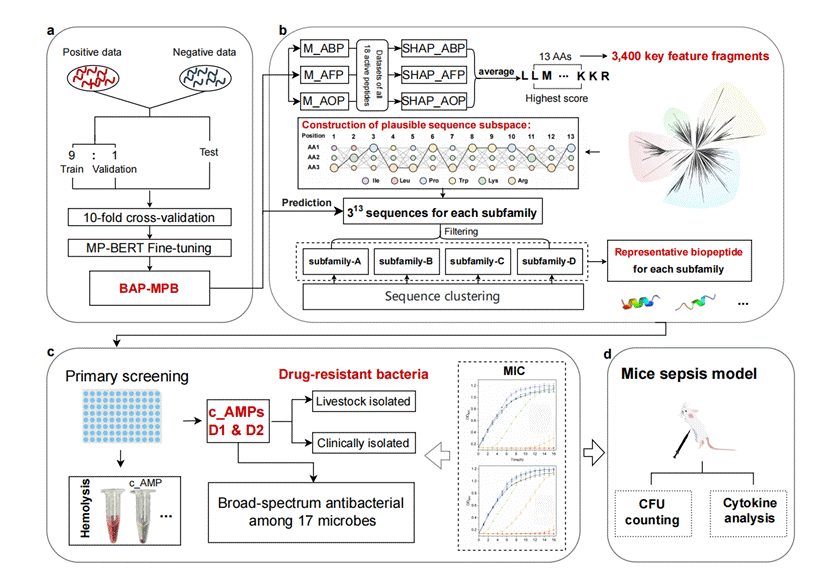
In this study, researchers utilized their self-developed protein large language model, MP-TRANS, to create a series of tools for predicting active peptides. This enabled an efficient transfer from general protein representation to AMP design. By incorporating SHAP-based interpretability analysis, they quantitatively assessed the contribution of each amino acid residue to antibacterial, antifungal, and antioxidant activities. The team identified high-contribution fragments and constructed a peptide sequence subspace for further testing.
Experimental validation demonstrated that out of 16 selected peptides, 12 exhibited two or more biological activities. Notably, the candidate peptide D1 displayed strong inhibitory effects against multidrug-resistant bacteria both in vitro and in vivo. This study represents a breakthrough in rational AMP design, combining “few-shot learning” with high precision, and highlights the critical role of protein large language models and domestic high-performance computing in advancing peptide research.
The study was supported by the National Key Research and Development Program of China, the National Natural Science Foundation of China, and the CAAS Science and Technology Innovation Project, with additional support from several collaborating institutions.
Article Link: https://www.nature.com/articles/s41467-025-64378-y

