
The milk research team of Institute of Animal Sciences, Chinese Academy of Agricultural Sciences systematically review the new methods of metagenome-based isolation and cultivation of uncultured microorganisms, which will contribute to achieve new breakthroughs in the isolation and cultivation of uncultured microorganisms. This study is published in the journal of Microbiome.
For the past few years, the rapid development of metagenomic sequencing technology has made researchers realize that there are vast uncultured microbial communities in various environment and they also play an important role in the Earth's ecosystem. Although metagenomic sequence have enhanced the understanding of the diversity and function of uncultured microbial communities, without the pure culture strains, it is not possible to directly assess the metabolic and physiological functions, nor to verify the interactions between microbial species. Therefore, the isolation and culture of uncultured microorganisms is of great significance for driving the research of microbiome into deep end, and for the development of biological or health industry as a new opportunity.
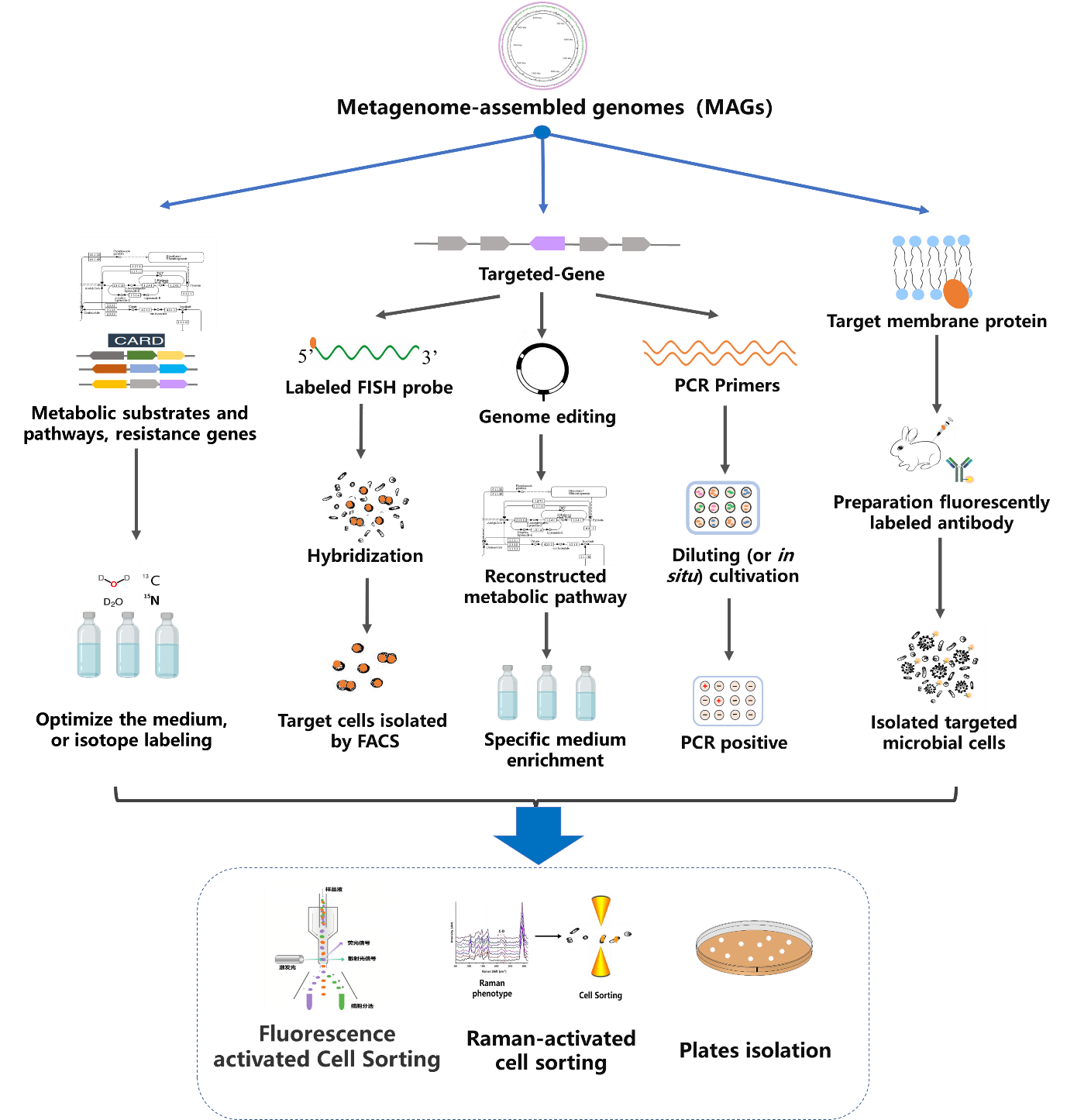
Isolating uncultured microorganisms must rely on new technologies and thoughts. The current explosion of metagenomic data, especially metagenomic assembly genome (MAG), provides genome information of uncultured microorganisms, which brings new opportunities for guiding the isolation and cultivation of target uncultured microorganisms.
By reviewing a great deal of literature and combining with their own research on uncultured microorganisms, the authors summarized and proposed methods for isolation and cultivation of uncultured microorganisms based on metagenomic data, including culture medium optimization, specific antibodies guided isolation, targeted functional genes screening and isolation. (1) Culture medium optimization. Metagenomic data provides the metabolism substrate, oxygen demand, and antibiotic resistance of targeted microbe. According to the information designing cultivation medium or growing conditions, combined with plates isolation or stable isotope labeling for the Raman-activated cell sorting methods to targeted isolation and cultivation microbe. (2) Specific antibodies guided isolation. Predicting the membrane proteins or epitopes by the target microbial genome, then preparing specific antibodies and labeled with fluorescence. The labeled antibodies are bound to the target microbes, and then isolated using Fluorescence activated Cell Sorting technology. (3) Targeted functional genes screening and isolation. Based on the target gene designed specific primers or probe, then screening by PCR or fluorescence in situ hybridization, combining with cell isolation methods to obtain target cells. Even more, the editing of antibiotics or substrates genes based on MAGs sequence information can facilitate the design of growth medium, which enables isolated targeted microbial cells.
The work was supported by Modern Agro-Industry Technology Research System of the PR China, and the Agricultural Science and Technology Innovation Program. The first author was Siji Liu. Corresponding authors were Shengguo Zhao and Jiaqi Wang.
The online version available at:https://doi.org/10.1186/s40168-022-01272-5

