
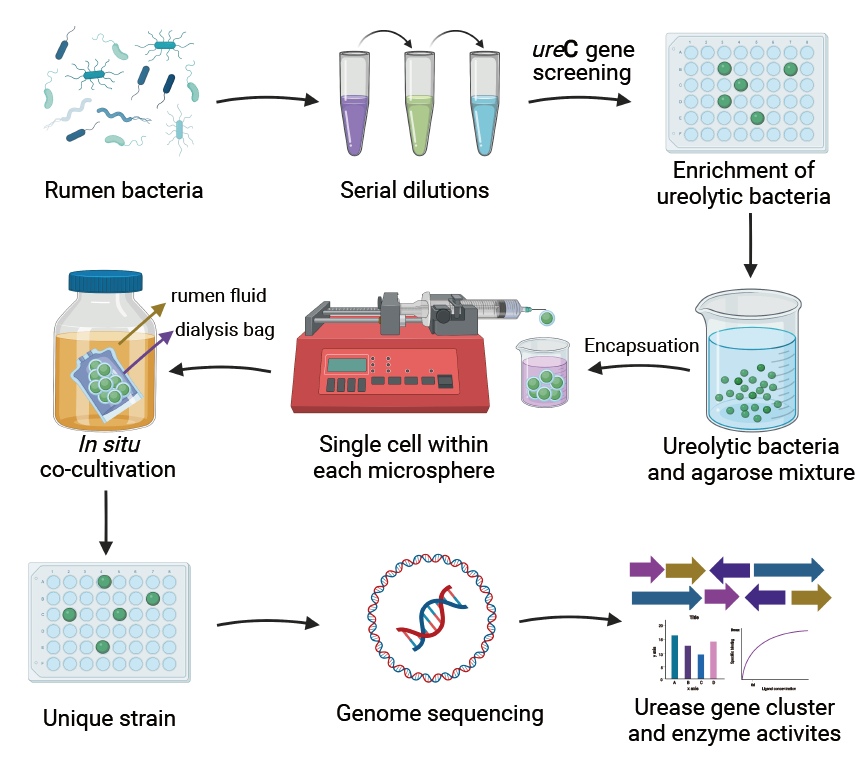
Milk Research Team of Institute of animal science of CAAS established a microbe isolation method which include target-gene enrichment plus in situ agarose microsphere cultivation. Crucial ureolytic bacteria with urease activity were isolated from the rumen using the targeted method, which enriched the rumen microbiome resources and will also help to better bridge the knowledge gap between genotypes and phenotypes of uncultured bacteria. The research was published in Microbiome.
Rumen ureolytic bacteria are the key microbes in regulating urea nitrogen utilization in ruminants. Sequencing-based studies have helped gain new insights into ruminal ureolytic bacterial diversity, but only a limited number of ureolytic bacteria have been isolated into pure cultures or studied, hindering the understanding of ureolytic bacteria with respect to their metabolism, physiology, and ecology.
For improve the efficiency of microbial isolation, established an integrated approach, which include urease gene guided enrichment plus in situ agarose microsphere (diameter 3.4mm) embedding and cultivation under rumen-simulating conditions. In total, we isolated 976 strains of bacteria, of which 52 strains were selected for genomic sequencing. Genomic analyses revealed that 28 strains, which were classifed into 12 species, contained urease genes, which indicated that the targeted isolation efficiency reached 54%. Compared to all the previously isolated ruminal ureolytic species combined, the newly isolated ureolytic bacteria increased the number of genotypically and phenotypically characterized ureolytic species by 34.38% and 45.83%, respectively. Pan-genomics analyses have shown that these isolated strains have unique genes compared to the known ureolytic strains of the same species indicating their new metabolic functions, especially in energy and nitrogen metabolism. All the ureolytic species were ubiquitous in the rumen of six different species of ruminants and were correlated to dietary urea metabolism in the rumen and milk protein production which indicated that the new strains had significant distribution and functional importance. Isolation strains had five different organizations of urease gene clusters, and the type of gene cluster uncorrelated with the ability to hydrolyze urea. The key amino acids residues that regulate urease activity in Flap region of active center were revealed through urease structure modeling.
This work was supported by the National Natural Science Foundation of China, Agricultural Science and Technology Innovation Program, Modern Agro-Industry Technology Research System of China, and State Key Laboratory of Animal Nutrition. First author is Sijia Liu, Corresponding author are Shengguo Zhao, Jiaqi Wang.
By Zhao Shengguo (zhaoshengguo@caas.cn)
Links go to the original paper: https://doi.org/10.1186/s40168-023-01510-4.

